Bayesian optimization of a Y branch#
Note: the cost of running the entire notebook is larger than 10 FlexCredits.
Bayesian optimization is a popular technique for searching and optimizing a design space. It works by building a probabilistic model, such as a Gaussian process, to predict the outcome of a more complex objective function. Unlike other optimizers like genetic algorithms or particle swarm optimization, Bayesian optimization uses an acquisition function to select new potential solutions, focusing on areas of the design space that the Gaussian process predicts will have high objective values and high uncertainty. Over the course of the optimization, the model’s uncertainty reduces, improving its accuracy as a surrogate for the true objective function. This makes Bayesian optimization a direct approach to optimization, particularly well-suited to problems with small (< 30 dimensions) and expensive-to-evaluate objective functions.
However, as Bayesian optimization is an iterative process with many individual components, it can be challenging to design and parallelize, leading to long run times and high computational costs. Fortunately, the Design plugin in Tidy3D includes the Bayesian optimization method MethodBayOpt which eliminates much of this complexity. Users can quickly and easily run a Bayesian optimization for complex FDTD simulations and analyze the results. Further details on Bayesian optimization and
the methods used in Tidy3D are available in the open-source Python library bayesian_optimization.
This notebook details how to perform a Bayesian optimization with the Tidy3D Design plugin for the development of a Y-junction. It is based on the work of Zhengqi Gao, Zhengxing Zhang, and Duane S. Boning, "Automatic Synthesis of Broadband Silicon Photonic Devices via Bayesian Optimization" J. Light. Technol. 40, 7879-7892 (2022) DOI:10.1109/JLT.2022.3207052. A detailed description of how to build a Y-junction can be found in the Waveguide
Y junction.
If you are curious about other Design plugin features, see these notebooks:
[1]:
# The Bayesian optimizer uses the bayesian-optimization external package version 1.5.1.
# Uncomment the following line to install the package
# pip install bayesian-optimization==1.5.1
import gdstk
import matplotlib.pyplot as plt
import numpy as np
import tidy3d as td
import tidy3d.plugins.design as tdd
import tidy3d.web as web
from scipy.interpolate import make_interp_spline
Simulation Setup#
The simulation is defined across the 1.5 \(\mu m\) to 1.6 \(\mu m\) wavelength range with 100 sampling points. The base of the model is silicon, the top of the model is silicon dioxide; we use the material constants included in the Tidy3D Tidy3D’s material library.
[2]:
lda0 = 1.55 # central wavelength
freq0 = td.C_0 / lda0 # central frequency
n_wav = 100 # Number of wavelengths to sample in the range
ldas = np.linspace(1.5, 1.6, n_wav) # wavelength range
freqs = td.C_0 / ldas # frequency range
fwidth = 0.5 * (np.max(freqs) - np.min(freqs)) # width of the source frequency range
si = td.material_library["cSi"]["Palik_Lossless"]
sio2 = td.material_library["SiO2"]["Palik_Lossless"]
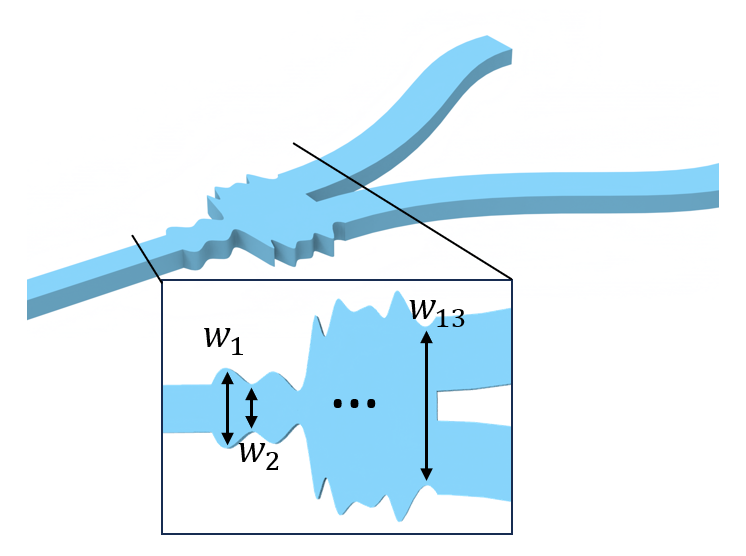
The following parameters describe the geometry of the waveguide. The optimizable design space is the junction which is split into 13 discrete segments.
[3]:
t = 0.22 # thickness of the silicon layer
num_d = 13 # dimensional space of the design region
l_in = 1 # input waveguide length
l_junction = 2 # length of the junction
l_bend = 6 # horizontal length of the waveguide bend
h_bend = 2 # vertical offset of the waveguide bend
l_out = 1 # output waveguide length
branch_width = 0.5 # width of one Y branch
branch_sep = 0.2 # distance between y branches at the junction
inf_eff = 100 # effective infinity
The most effective way to use the Design plugin is to split the workflow into “pre” and “post” functions which can surround a call to the Tidy3D cloud that carries out the computation. This takes advantage of automated simulation batching, allowing for parallelization which saves a considerable amount of time. The pre-function returns a Simulation; the post-function then analyzes the corresponding SimulationData. Together they can be considered the fitness function (or objective function
/ figure of merit) which the Bayesian optimization process is working to predict.
The function fn_pre needs to take the parameters that are being optimized: these are the 13 width segments of the junction. They always input as a dictionary and can be unpacked within the function (as below) or included as keyword arguments. These parameters are used to build a Y-junction Simulation object.
The function fn_post then takes the SimulationData object output by the simulation and computes the objective function. The value of objective is then fed back into the Bayesian optimization to inform the probabilistic model. In this case, we extract the power passing through the Y-junction and the power reflected back to the source. These values are evaluated in a custom loss function described by Gao et al. to determine the effectiveness of the junction design:
end{equation}` where the summation is performed over the simulated wavelengths, \(N_\lambda\) is the number of wavelength points, \(R\) is the reflected power and \(T\) is the transmitted power in one branch.
The aim of this function is to achieve a transmitted power of 0.5 through each branch, whilst driving the power reflected towards the source to zero. Note there is a minus sign in front of the output value as this loss function is a minimizing function, whilst all the optimizers in the Design plugin are built to maximize the objective function.
[4]:
def fn_pre(**w_params: dict) -> td.Simulation:
"""Create a Simulation of a Y splitter from a series of junction widths.
Includes mode monitors to measure the power transmitted and reflected to source.
"""
w_start = 0.5
w_end = branch_width * 2 + branch_sep
widths = [w_start] # Ensures input waveguide is included in spline for first point of junction
widths.extend(list(w_params.values()))
widths.append(w_end) # Ensures final point of junction smoothly converts to the branches
x_junction = np.linspace(
l_in, l_in + l_junction, num_d + 2
) # x coordinates of the top edge vertices
y_junction = np.array(widths) # y coordinates of the top edge vertices
# pass vertices through spline and increase sampling to smooth the geometry
new_x_junction = np.linspace(
l_in, l_in + l_junction, 100
) # x coordinates of the top edge vertices
spline = make_interp_spline(x_junction, y_junction, k=2)
spline_yjunction = spline(new_x_junction)
# using concatenate to include bottom edge vertices
x_junction = np.concatenate((new_x_junction, np.flipud(new_x_junction)))
y_junction = np.concatenate((spline_yjunction / 2, -np.flipud(spline_yjunction / 2)))
# stacking x and y coordinates to form vertices pairs
vertices = np.transpose(np.vstack((x_junction, y_junction)))
junction = td.Structure(
geometry=td.PolySlab(vertices=vertices, axis=2, slab_bounds=(0, t)), medium=si
)
x_start = l_in + l_junction # x coordinate of the starting point of the waveguide bends
x_bend = np.linspace(x_start, x_start + l_bend, 100) # x coordinates of the top edge vertices
y_bend = (
(x_bend - x_start) * h_bend / l_bend
- h_bend * np.sin(2 * np.pi * (x_bend - x_start) / l_bend) / (np.pi * 2)
+ w_end / 2
- w_start / 2
) # y coordinates of the top edge vertices
# adding the last point to include the straight waveguide at the output
x_bend = np.append(x_bend, inf_eff)
y_bend = np.append(y_bend, y_bend[-1])
# add path to the cell
cell = gdstk.Cell("bends")
cell.add(
gdstk.FlexPath(x_bend + 1j * y_bend, branch_width, layer=1, datatype=0)
) # top waveguide bend
cell.add(
gdstk.FlexPath(x_bend - 1j * y_bend, branch_width, layer=1, datatype=0)
) # bottom waveguide bend
# define top waveguide bend structure
wg_bend_1 = td.Structure(
geometry=td.PolySlab.from_gds(
cell,
gds_layer=1,
axis=2,
slab_bounds=(0, t),
)[0],
medium=si,
)
# define bottom waveguide bend structure
wg_bend_2 = td.Structure(
geometry=td.PolySlab.from_gds(
cell,
gds_layer=1,
axis=2,
slab_bounds=(0, t),
)[1],
medium=si,
)
# straight input waveguide
wg_in = td.Structure(
geometry=td.Box.from_bounds(rmin=(-inf_eff, -w_start / 2, 0), rmax=(l_in, w_start / 2, t)),
medium=si,
)
# the entire model is the collection of all structures defined so far
model_structure = [wg_in, junction, wg_bend_1, wg_bend_2]
Lx = l_in + l_junction + l_out + l_bend # simulation domain size in x direction
Ly = w_end + 2 * h_bend + 1.5 * lda0 # simulation domain size in y direction
Lz = 10 * t # simulation domain size in z direction
sim_size = (Lx, Ly, Lz)
# add a mode source as excitation
mode_spec = td.ModeSpec(num_modes=1, target_neff=3.5)
mode_source = td.ModeSource(
center=(l_in / 2, 0, t / 2),
size=(0, 4 * w_start, 6 * t),
source_time=td.GaussianPulse(freq0=freq0, fwidth=fwidth),
direction="+",
mode_spec=mode_spec,
mode_index=0,
)
# add a mode monitor to measure transmission at the output waveguide
mode_monitor_11 = td.ModeMonitor(
center=(l_in / 3, 0, t / 2),
size=(0, 4 * w_start, 6 * t),
freqs=freqs,
mode_spec=mode_spec,
name="mode_11",
)
mode_monitor_12 = td.ModeMonitor(
center=(l_in + l_junction + l_bend + l_out / 2, w_end / 2 - w_start / 2 + h_bend, t / 2),
size=(0, 4 * w_start, 6 * t),
freqs=freqs,
mode_spec=mode_spec,
name="mode_12",
)
# add a field monitor to visualize field distribution at z=t/2
field_monitor = td.FieldMonitor(
center=(0, 0, t / 2), size=(td.inf, td.inf, 0), freqs=[freq0], name="field"
)
run_time = 5e-13 # simulation run time
# construct simulation
sim = td.Simulation(
center=(Lx / 2, 0, 0),
size=sim_size,
grid_spec=td.GridSpec.auto(min_steps_per_wvl=20, wavelength=lda0),
structures=model_structure,
sources=[mode_source],
monitors=[mode_monitor_11, mode_monitor_12, field_monitor],
run_time=run_time,
boundary_spec=td.BoundarySpec.all_sides(boundary=td.PML()),
medium=sio2,
)
return sim
def fn_post(sim_data: td.SimulationData) -> float:
"""Calculate the loss function from the power at the mode monitors in the SimulationData."""
# Calculate the power reflected back to source and transmitted to one branch
power_reflected = np.squeeze(
np.abs(sim_data["mode_11"].amps.sel(direction="-", mode_index=0)) ** 2
)
power_transmitted = np.squeeze(
np.abs(sim_data["mode_12"].amps.sel(direction="+", mode_index=0)) ** 2
)
# Loss function proposed by Gao et al. which takes advantage of branch symmetry
loss_fn = 1 / 3 * n_wav * np.sum(power_reflected**2 + 2 * (power_transmitted - 0.5) ** 2)
output = -float(loss_fn.values) # Negative value as this is a minimizing loss function
return output
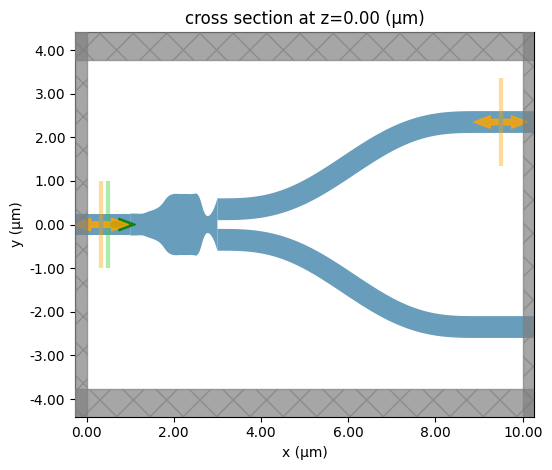
We can quickly check that fn_pre is working correctly by passing a set of potential test_params and plotting the result. Note that the gap between the Polyslab junction and the branches is a plotting artifact and doesn’t exist in the Simulation.
[5]:
test_params = {
"w1": 0.5,
"w2": 0.5,
"w3": 0.6,
"w4": 0.7,
"w5": 0.9,
"w6": 1.26,
"w7": 1.4,
"w8": 1.4,
"w9": 1.4,
"w10": 1.4,
"w11": 1.31,
"w12": 0.5,
"w13": 0.5,
}
sim = fn_pre(**test_params)
sim.plot(z=0)
plt.show()
Next, we setup the Bayesian optimization method. This is done with the MethodBayOpt object. We don’t have the lcb (lower confidence bound) acquisition function used in the paper available to us, but by making our loss function negative and using the ucb (upper confidence bound) acquisition we achieve the same optimizer design. The initial_iter and n_iter options control the number of initial random samples and subsequent optimization iterations respectively. We can also set
the random seed to ensure reproducible results (optional).
We also create a list of ParameterFloat objects corresponding to the 13 segment widths. The span option defines the bounds of each parameter.
The method and parameters are then passed to a DesignSpace object which contains all we need to run the Bayesian optimization.
[6]:
method = tdd.MethodBayOpt(
initial_iter=30,
n_iter=70,
acq_func="ucb",
kappa=0.3,
seed=1,
)
parameters = [tdd.ParameterFloat(name=f"w_{i}", span=(0.5, 1.6)) for i in range(num_d)]
design_space = tdd.DesignSpace(
method=method, parameters=parameters, task_name="bay_opt_notebook", path_dir="./data"
)
It is then very easy to the launch this optimization with design_space.run(). This launches an initial random batch of 30 simulations, as specified by initial_iter, followed by sequential computation of 70 simulations, as specified by n_iter. In the latter phase, the Bayesian optimizer chooses potential candidates based on simultaneously maximizing the objective value and efficiently exploring the design space. The total of 100 simulations takes around 90 minutes to compute. Once
complete, the results are returned in a pandas dataframe for analysis.
[7]:
results = design_space.run(fn_pre, fn_post, verbose=True)
df = results.to_dataframe()
22:01:49 CEST Running 30 Simulations
22:03:54 CEST Best Fit from Initial Solutions: -181.226
Running 1 Simulations
22:04:03 CEST Running 1 Simulations
22:04:13 CEST Running 1 Simulations
22:04:21 CEST Latest Best Fit on Iter 2: -150.359
22:04:22 CEST Running 1 Simulations
22:04:30 CEST Latest Best Fit on Iter 3: -134.853
22:04:31 CEST Running 1 Simulations
22:04:40 CEST Latest Best Fit on Iter 4: -122.553
Running 1 Simulations
22:04:49 CEST Running 1 Simulations
22:04:58 CEST Latest Best Fit on Iter 6: -81.173
Running 1 Simulations
22:05:08 CEST Running 1 Simulations
22:05:17 CEST Running 1 Simulations
22:05:26 CEST Running 1 Simulations
22:05:35 CEST Running 1 Simulations
22:05:44 CEST Latest Best Fit on Iter 11: -68.01
Running 1 Simulations
22:05:53 CEST Running 1 Simulations
22:06:03 CEST Running 1 Simulations
22:06:12 CEST Running 1 Simulations
22:06:21 CEST Running 1 Simulations
22:06:30 CEST Latest Best Fit on Iter 16: -44.9
Running 1 Simulations
22:06:40 CEST Running 1 Simulations
22:06:49 CEST Running 1 Simulations
22:06:58 CEST Latest Best Fit on Iter 19: -35.059
Running 1 Simulations
22:07:07 CEST Latest Best Fit on Iter 20: -27.09
Running 1 Simulations
22:07:16 CEST Latest Best Fit on Iter 21: -23.111
Running 1 Simulations
22:07:25 CEST Latest Best Fit on Iter 22: -19.258
22:07:26 CEST Running 1 Simulations
22:07:35 CEST Running 1 Simulations
22:07:44 CEST Latest Best Fit on Iter 24: -15.179
Running 1 Simulations
22:07:53 CEST Running 1 Simulations
22:08:02 CEST Running 1 Simulations
22:08:12 CEST Running 1 Simulations
22:08:21 CEST Running 1 Simulations
22:08:30 CEST Latest Best Fit on Iter 29: -12.722
Running 1 Simulations
22:08:39 CEST Running 1 Simulations
22:08:48 CEST Latest Best Fit on Iter 31: -12.159
Running 1 Simulations
22:08:57 CEST Running 1 Simulations
22:09:06 CEST Latest Best Fit on Iter 33: -11.087
22:09:07 CEST Running 1 Simulations
22:09:15 CEST Latest Best Fit on Iter 34: -10.551
22:09:16 CEST Running 1 Simulations
22:09:25 CEST Running 1 Simulations
22:09:34 CEST Latest Best Fit on Iter 36: -7.42
Running 1 Simulations
22:09:43 CEST Running 1 Simulations
22:09:52 CEST Latest Best Fit on Iter 38: -6.507
22:09:53 CEST Running 1 Simulations
22:10:02 CEST Running 1 Simulations
22:10:11 CEST Running 1 Simulations
22:10:21 CEST Running 1 Simulations
22:10:30 CEST Latest Best Fit on Iter 42: -6.414
Running 1 Simulations
22:10:39 CEST Running 1 Simulations
22:10:48 CEST Running 1 Simulations
22:10:57 CEST Latest Best Fit on Iter 45: -5.994
22:10:58 CEST Running 1 Simulations
22:13:06 CEST Running 1 Simulations
22:13:53 CEST Running 1 Simulations
22:14:38 CEST Running 1 Simulations
22:15:20 CEST Running 1 Simulations
22:16:03 CEST Latest Best Fit on Iter 50: -5.513
Running 1 Simulations
22:16:46 CEST Running 1 Simulations
22:18:12 CEST Running 1 Simulations
22:19:32 CEST Running 1 Simulations
22:20:15 CEST Running 1 Simulations
22:21:04 CEST Latest Best Fit on Iter 55: -5.463
Running 1 Simulations
22:21:53 CEST Latest Best Fit on Iter 56: -5.229
Running 1 Simulations
22:22:42 CEST Latest Best Fit on Iter 57: -4.891
Running 1 Simulations
22:23:26 CEST Latest Best Fit on Iter 58: -4.182
22:23:27 CEST Running 1 Simulations
22:24:12 CEST Running 1 Simulations
22:24:56 CEST Latest Best Fit on Iter 60: -3.962
Running 1 Simulations
22:25:41 CEST Running 1 Simulations
22:27:10 CEST Running 1 Simulations
22:27:56 CEST Latest Best Fit on Iter 63: -3.735
Running 1 Simulations
22:28:41 CEST Running 1 Simulations
22:29:25 CEST Latest Best Fit on Iter 65: -3.544
22:29:26 CEST Running 1 Simulations
22:30:17 CEST Latest Best Fit on Iter 66: -3.42
Running 1 Simulations
22:31:01 CEST Latest Best Fit on Iter 67: -3.288
22:31:02 CEST Running 1 Simulations
22:31:53 CEST Running 1 Simulations
22:33:28 CEST Latest Best Fit on Iter 69: -3.256
Best Result: -3.255581414447776 Best Parameters: w_0: 0.5188415764684946 w_1: 0.5 w_10: 0.5 w_11: 0.5 w_12: 1.1177014993031638 w_2: 1.1415176565004286 w_3: 1.6 w_4: 1.2733923550648545 w_5: 1.3868594525894349 w_6: 1.419242614773869 w_7: 1.2066095530458796 w_8: 1.6 w_9: 1.1296647155637014
Results#
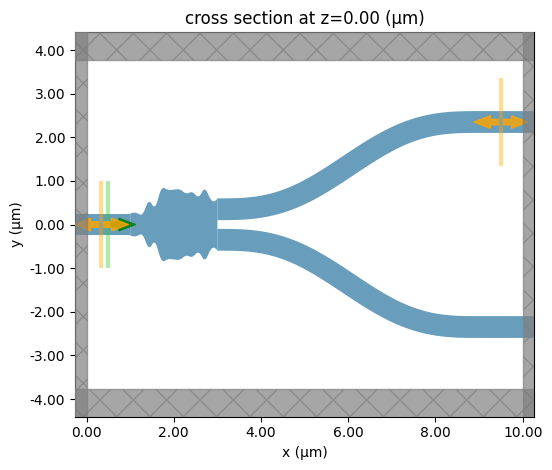
The best result can be extracted directly from the optimizer object. Plotting this, we see what the optimizer has returned as the optimized structure for this design of Y-junction.
[8]:
best_params = results.optimizer.max["params"]
print(f"Best fitness: {results.optimizer.max['target']}")
sim = fn_pre(**best_params)
sim.plot(z=0)
plt.show()
Best fitness: -3.255581414447776
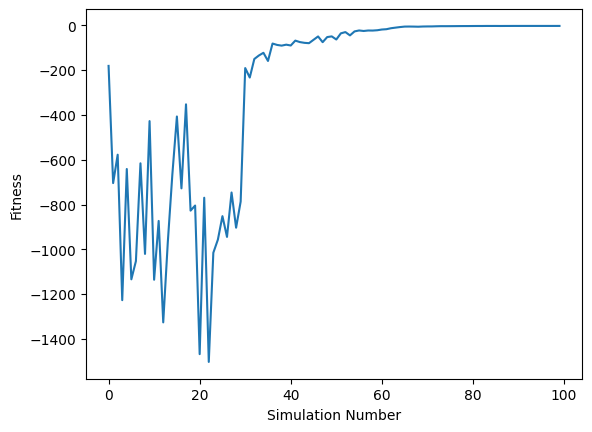
We can then create a plot to evaluate if the Bayesian optimization has converged on a fitness value. The first 30 iterations are from the random initialization, so the fitness values are expected to be more varied. The fitness has converged before the end of the remaining 70 iterations; an early stop criteria could have been included to finish the optimization sooner.
[9]:
ax = df["output"].plot(xlabel="Simulation Number", ylabel="Fitness")
The mean power at the reflected and transmitted monitors can be calculated and compared to the paper from Gao et al.
[10]:
idx_best_result = df["output"].idxmax()
best_sim_filename = results.task_paths[idx_best_result]
best_sim = td.SimulationData.from_file(best_sim_filename)
power_reflected = np.array(
np.squeeze(np.abs(best_sim["mode_11"].amps.sel(direction="-", mode_index=0)) ** 2)
).mean()
power_transmitted = np.array(
np.squeeze(np.abs(best_sim["mode_12"].amps.sel(direction="+", mode_index=0)) ** 2)
).mean()
print(f"Mean Reflected Power: {round(power_reflected, 3)} (Paper: 0.004)")
print(f"Mean Transmitted Power: {round(power_transmitted, 3)} (Paper: 0.460)")
Mean Reflected Power: 0.001 (Paper: 0.004)
Mean Transmitted Power: 0.478 (Paper: 0.460)
Final Optimized Design#
Finally, we can simulate the optimal solution with an additional FieldMonitor to visualise the field distribution throughout the splitter.
[11]:
final_sim = fn_pre(**best_params)
# Define a field monitor to help visualize the field distribution
field_monitor = td.FieldMonitor(
center=(0, 0, 0), size=(td.inf, td.inf, 0), freqs=[freq0], name="field"
)
mode_12 = final_sim.monitors[1]
final_sim = final_sim.copy(update={"monitors": (field_monitor, mode_12)})
final_sim_data = web.run(final_sim, task_name="BO_notebook_final_sim")
Created task 'BO_notebook_final_sim' with task_id 'fdve-1c9aafc2-a589-4866-aa95-880853947d87' and task_type 'FDTD'.
22:33:29 CEST View task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-1c9aafc2-a5 89-4866-aa95-880853947d87'.
Task folder: 'default'.
22:33:30 CEST Maximum FlexCredit cost: 0.111. Minimum cost depends on task execution details. Use 'web.real_cost(task_id)' to get the billed FlexCredit cost after a simulation run.
22:33:31 CEST status = queued
To cancel the simulation, use 'web.abort(task_id)' or 'web.delete(task_id)' or abort/delete the task in the web UI. Terminating the Python script will not stop the job running on the cloud.
22:34:45 CEST status = preprocess
22:34:49 CEST starting up solver
running solver
22:35:11 CEST early shutoff detected at 76%, exiting.
22:35:12 CEST status = postprocess
22:35:14 CEST status = success
22:35:16 CEST View simulation result at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-1c9aafc2-a5 89-4866-aa95-880853947d87'.
22:35:20 CEST loading simulation from simulation_data.hdf5
Plotting the field shows how it splits evenly between each branch.
[12]:
final_sim_data.plot_field("field", "E", "abs^2")
plt.show()
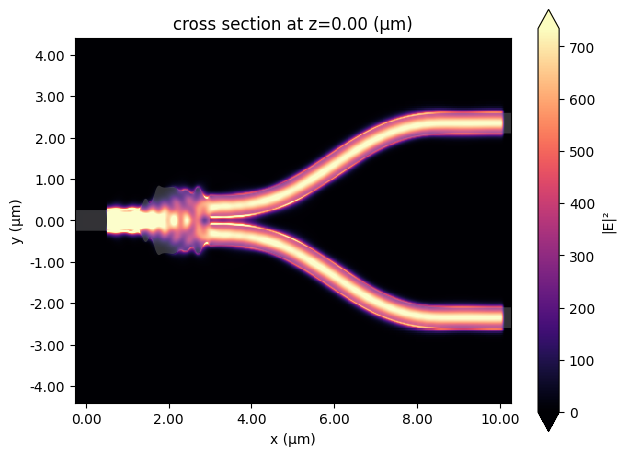
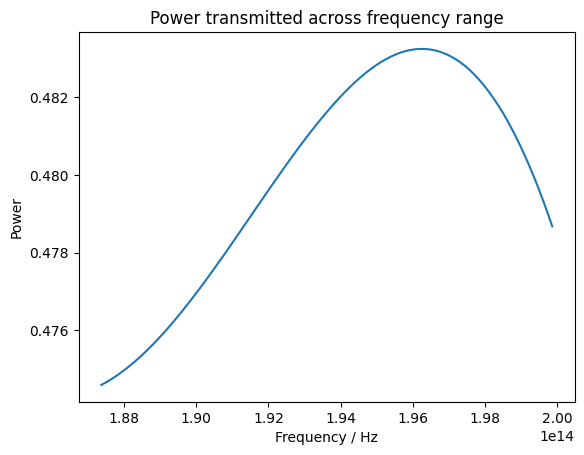
And we can visualise how the power transmitted varies over the frequency range.
[13]:
power_transmitted = np.squeeze(
np.abs(final_sim_data["mode_12"].amps.sel(direction="+", mode_index=0)) ** 2
)
plt.plot(freqs, power_transmitted)
plt.title("Power transmitted across frequency range")
plt.xlabel("Frequency / Hz")
plt.ylabel("Power")
plt.show()
Conclusion#
Through comparison of the junction geometry and the transmitted and reflected power, we can show that these results closely follow the results published by Gao et al. for the design of a Y-junction. This notebook demonstrates how to carry out Bayesian optimization with the Design plugin, and can be readily adapted to other use cases.
Learn more about the Design plugin: