Waveguide Crossing¶
This example uses of PhotonForge to optimize a waveguide crossing based on [1]. A Monte Carlo parameter sweep is used to explore the initial design space and provide a good optimization starting point. The local optimization uses the Nelder-Mead method based on scipy’s minimize function.
References
Sujith Chandran, Marcus Dahlem, Yusheng Bian, Paulo Moreira, Ajey P. Jacob, Michal Rakowski, Andy Stricker, Karen Nummy, Colleen Meagher, Bo Peng, Abu Thomas, Shuren Hu, Jan Petykiewicz, Zoey Sowinski, Won Suk Lee, Rod Augur, Dave Riggs, Ted Letavic, Anthony Yu, Ken Giewont, John Pellerin, and Jaime Viegas, “Beam shaping for ultra-compact waveguide crossings on monolithic silicon photonics platform,” Opt. Lett. 45, 6230-6233 (2020), doi: 10.1364/OL.402446
This example has a similar Tidy3D version: Waveguide crossing based on cosine tapers.
[1]:
import numpy as np
import photonforge as pf
import scipy.optimize
import tidy3d as td
from matplotlib import pyplot as plt
For this demonstration, the basic_technology is enough to show the main concepts behind it. Other technologies can be equally used to optimize the crossing for other PDKs.
[2]:
tech = pf.basic_technology()
pf.config.default_technology = tech
Geometry Parametrization¶
PhotonForge provides a flexible crossing parametric component that can be directly used to re-create the cosine-tapered geometry proposed in the main reference.
We create a custom function with the parameters we’re interested in optimizing as variables. Although possible, we don’t really need to make this function a parametric component function only to optimize those parameters, considering we already return a parametric crossing from it.
Besides the re-parametrization, we also set the Tidy3D model’s arguments to remove verbosity and increase the default run time because, depending on the exact parameters, the geometry can become resonant.
[3]:
def sine_crossing(amplitude, arm_length):
model = pf.Tidy3DModel(verbose=False, run_time=3e-12)
crossing = pf.parametric.crossing(
port_spec="Strip",
arm_length=arm_length,
extra_length=0.5,
added_width=f"{amplitude} * sin(pi * u)",
model=model,
)
return crossing
sine_crossing(arm_length=3, amplitude=1)
[3]:
Monte Carlo Exploration¶
The initial exploration of the parameter space is done with the help of the Monte Carlo tools in PhotonForge. We can calculate a range of S parameters for variations of the crossing by setting its parameters as random variables within a fixed value range. The ranges are quite wide because we know almost nothing a priori about the design, therefore the number of samples is also considerably high to allow proper exploration of the whole space.
Note that because we are only interested in the transmission from P0 to P2, we explicitly set the ports and modes we want to run as sources for the Tidy3D simulations using the inputs argument.
[4]:
freqs = pf.C_0 / np.linspace(1.45, 1.65, 21)
variables, results = pf.monte_carlo.s_matrix(
sine_crossing,
freqs,
("amplitude", "component", {"value_range": (0.1, 1.5)}),
("arm_length", "component", {"value_range": (0.5, 3)}),
random_samples=30,
component_kwargs={"amplitude": 1, "arm_length": 3},
model_kwargs={"inputs": ["P0@0"]},
random_seed=0,
)
Starting sample 1 of 30…
Starting sample 2 of 30…
Starting sample 3 of 30…
Starting sample 4 of 30…
Starting sample 5 of 30…
Starting sample 6 of 30…
Starting sample 7 of 30…
Starting sample 8 of 30…
Starting sample 9 of 30…
Starting sample 10 of 30…
Starting sample 11 of 30…
Starting sample 12 of 30…
Starting sample 13 of 30…
Starting sample 14 of 30…
Starting sample 15 of 30…
Starting sample 16 of 30…
Starting sample 17 of 30…
Starting sample 18 of 30…
Starting sample 19 of 30…
Starting sample 20 of 30…
Starting sample 21 of 30…
Starting sample 22 of 30…
Starting sample 23 of 30…
Starting sample 24 of 30…
Starting sample 25 of 30…
Starting sample 26 of 30…
Starting sample 27 of 30…
Starting sample 28 of 30…
Starting sample 29 of 30…
Starting sample 30 of 30…
Sample 1 done.
Sample 2 done.
Sample 3 done.
Sample 4 done.
Sample 5 done.
Sample 6 done.
Sample 7 done.
Sample 8 done.
Sample 9 done.
Sample 10 done.
Sample 11 done.
Sample 12 done.
Sample 13 done.
Sample 14 done.
Sample 15 done.
Sample 16 done.
Sample 17 done.
Sample 18 done.
Sample 19 done.
Sample 20 done.
Sample 21 done.
Sample 22 done.
Sample 23 done.
Sample 24 done.
Sample 25 done.
Sample 26 done.
Sample 27 done.
Sample 28 done.
Sample 29 done.
Sample 30 done.
All samples done!
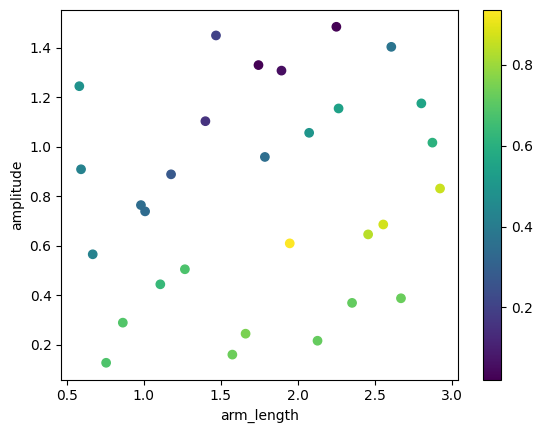
The results include the full S matrix for each crossing variation. To help us make sense of all this data, we can define a figure of merit for the design and plot it versus the crossing parameters. In this case, a reasonable definition is to use the minimal transmission value within the frequency range we’re interested in. Let’s first create a result table with only the parameters and our figure of merit, instead of the whole S matrix objects:
[5]:
names = [v.name for v in variables]
table = np.array([(*v, np.abs(s["P0@0", "P2@0"]).min() ** 2) for *v, s in results])
table
[5]:
array([[1.89293245, 1.30670413, 0.05278403],
[1.78530457, 0.95793781, 0.33876378],
[1.57431442, 0.15939063, 0.73916431],
[1.39931042, 1.10239893, 0.16325344],
[1.74379972, 1.32907849, 0.02224897],
[0.66835153, 0.56466724, 0.43891358],
[2.07250101, 1.05543504, 0.49373283],
[2.45512328, 0.64501438, 0.84189003],
[2.55332675, 0.68507742, 0.871529 ],
[2.12682069, 0.21543469, 0.7157571 ],
[0.98118614, 0.76343902, 0.35743158],
[2.66823248, 0.38698483, 0.7140557 ],
[1.46807783, 1.44878793, 0.21607665],
[1.94662566, 0.60877627, 0.9339298 ],
[2.92168546, 0.83047987, 0.85811127],
[2.35086521, 0.36836535, 0.71804983],
[0.86364256, 0.28845985, 0.69798256],
[2.24903115, 1.48379688, 0.0237723 ],
[2.26344246, 1.15381968, 0.53899655],
[2.87219256, 1.0157638 , 0.59670117],
[0.59253126, 0.90813608, 0.44008817],
[1.26638186, 0.50425125, 0.67607769],
[1.00809177, 0.73810412, 0.34707558],
[0.58137162, 1.24378247, 0.49505836],
[2.80040379, 1.17426609, 0.55566748],
[1.10690058, 0.44344074, 0.64200541],
[1.17656064, 0.88775554, 0.28828981],
[0.75534563, 0.12637548, 0.68734295],
[2.60544912, 1.4027804 , 0.36232749],
[1.66009647, 0.24387858, 0.74917659]])
[6]:
plt.scatter(table[:, 0], table[:, 1], c=table[:, 2])
plt.xlabel(names[0])
plt.ylabel(names[1])
_ = plt.colorbar()
Local Optimization¶
We use scipy to optimize the crossing around the best result we found through the Nelder-Mead method, which does not require a gradient computation. More advanced optimization methods based on ajoint computations are available in Tidy3D directly, if the device require those due to, for example, a large number of parameters.
The only step required to run the optimization is the creation of an objective function to be minimized. It receives arguments as an array, which we decompose into the 2 parameters for our crossing. Furthermore, because we desire to maximize the minimal transmission, we use a negative sign in our figure of merit:
[7]:
def objective(args):
kwargs = dict(zip(names, args))
c = sine_crossing(**kwargs)
s_matrix = c.s_matrix(freqs)
min_transmission = np.abs(s_matrix["P0@0", "P2@0"]).min() ** 2
# We can print the current evaluation result to keep track of the optimization process
print(args, "→", min_transmission, flush=True)
return -min_transmission
For the actual optimization run, we limit the convergence tolerances to reasonable values, as well as the number of function evaluations so that we don’t run unnecessary simulations. If those values end up being too small, we can always restart the optimization from the last best parameters.
[8]:
best_variation = np.argmax(table[:, 2])
initial_guess = (table[best_variation, 0], table[best_variation, 1])
opt_result = scipy.optimize.minimize(
objective,
initial_guess,
method="Nelder-Mead",
bounds=[v.value_spec["value_range"] for v in variables],
options={"maxfev": 16, "fatol": 0.005, "xatol": 0.005},
)
opt_result
Progress: 100%
[1.94662566 0.60877627] → 0.9339298047399855
Progress: 100%
[2.04395694 0.60877627] → 0.9343944562269577
Progress: 100%
[1.94662566 0.63921508] → 0.9019294369173531
Progress: 100%
[2.04395694 0.57833745] → 0.9205140800772739
Progress: 100%
[2.01962412 0.59355686] → 0.9324986020857798
Progress: 100%
[1.97095848 0.62399567] → 0.919908097128029
Starting…
11:44:50 EST WARNING: Warning messages were found in the solver log. For more information, check 'SimulationData.log' or use 'web.download_log(task_id)'.
WARNING: Warning messages were found in the solver log. For more information, check 'SimulationData.log' or use 'web.download_log(task_id)'.
Progress: 100%
[2.00745771 0.60116656] → 0.936600464156625
Progress: 100%
[2.10478899 0.60116656] → 0.9216268648708215
Progress: 100%
[1.98616649 0.60687384] → 0.9396004402613354
Progress: 100%
[1.94966726 0.59926414] → 0.9365647424038442
Progress: 100%
[1.97323968 0.60164217] → 0.9380691968705966
Progress: 100%
[1.95194846 0.60734945] → 0.9345695174499332
Progress: 100%
[1.9935804 0.60271228] → 0.9382510058978215
Progress: 100%
[2.00650721 0.60794396] → 0.9391427175336075
Progress: 100%
[1.9990933 0.61210551] → 0.9369445609434127
Progress: 100%
[1.99495862 0.60506059] → 0.9390632398801809
[8]:
message: Maximum number of function evaluations has been exceeded.
success: False
status: 1
fun: -0.9396004402613354
x: [ 1.986e+00 6.069e-01]
nit: 8
nfev: 16
final_simplex: (array([[ 1.986e+00, 6.069e-01],
[ 2.007e+00, 6.079e-01],
[ 1.995e+00, 6.051e-01]]), array([-9.396e-01, -9.391e-01, -9.391e-01]))
Inspecting the Optimized Component¶
We can recreate the best variation for out technology and reset its model to remove the helper symmetries and include a field monitor if we want to fully inspect the component. Of course, with our parameters being layout dimensions, we should not use an arbitrary number of decimals, so we round them appropriately: in this case, 2 decimals (10 nm grid) should suffice.
[9]:
kwargs = dict(zip(names, np.round(opt_result.x, decimals=2)))
crossing = sine_crossing(**kwargs)
tidy3d_model = pf.Tidy3DModel(
monitors=[
td.FieldMonitor(
name="field",
center=(0, 0, 0.11),
size=(td.inf, td.inf, 0),
freqs=[freqs.mean()],
)
]
)
crossing.add_model(tidy3d_model, "Tidy3D")
s_matrix = crossing.s_matrix(freqs)
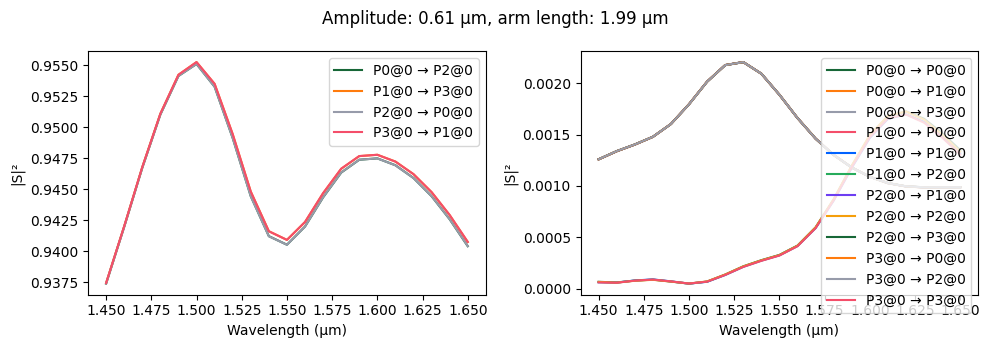
fig, ax = pf.plot_s_matrix(s_matrix)
_ = fig.suptitle(
f"Amplitude: {kwargs['amplitude']} μm, arm length: {kwargs['arm_length']} μm"
)
Starting…
11:44:55 EST Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-99dc8ac3-42e 7-4628-a59e-b41b24cf0be7'.
11:44:56 EST Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-18a1940d-fad a-4f11-9adc-be17205e763c'.
Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-50e276d0-e58 a-41fd-bf2b-44d519d9efe5'.
Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-f446629b-77a 3-48c1-9b8b-3b61aaa033a6'.
Progress: 100%
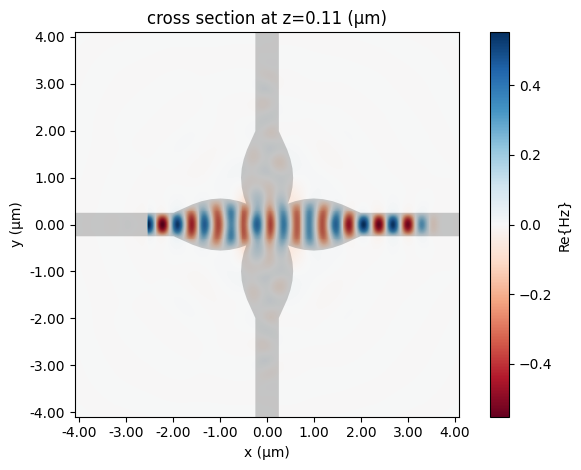
In order to plot the fields, we can use the batch_data function from the Tidy3D model to get the BatchData that includes the fields from our extra monitor:
[10]:
sim_data = tidy3d_model.batch_data(crossing, freqs)
_ = sim_data["P0@0"].plot_field("field", "Hz", robust=False)
Progress: 100%
11:44:57 EST Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-99dc8ac3-42e 7-4628-a59e-b41b24cf0be7'.
Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-18a1940d-fad a-4f11-9adc-be17205e763c'.
Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-50e276d0-e58 a-41fd-bf2b-44d519d9efe5'.
Loading simulation from local cache. View cached task using web UI at 'https://tidy3d.simulation.cloud/workbench?taskId=fdve-f446629b-77a 3-48c1-9b8b-3b61aaa033a6'.